Zongwei Zhou
Early and Prediagnostic Detection of Pancreatic Cancer from Computed Tomography
Jan 29, 2026Abstract:Pancreatic ductal adenocarcinoma (PDAC), one of the deadliest solid malignancies, is often detected at a late and inoperable stage. Retrospective reviews of prediagnostic CT scans, when conducted by expert radiologists aware that the patient later developed PDAC, frequently reveal lesions that were previously overlooked. To help detecting these lesions earlier, we developed an automated system named ePAI (early Pancreatic cancer detection with Artificial Intelligence). It was trained on data from 1,598 patients from a single medical center. In the internal test involving 1,009 patients, ePAI achieved an area under the receiver operating characteristic curve (AUC) of 0.939-0.999, a sensitivity of 95.3%, and a specificity of 98.7% for detecting small PDAC less than 2 cm in diameter, precisely localizing PDAC as small as 2 mm. In an external test involving 7,158 patients across 6 centers, ePAI achieved an AUC of 0.918-0.945, a sensitivity of 91.5%, and a specificity of 88.0%, precisely localizing PDAC as small as 5 mm. Importantly, ePAI detected PDACs on prediagnostic CT scans obtained 3 to 36 months before clinical diagnosis that had originally been overlooked by radiologists. It successfully detected and localized PDACs in 75 of 159 patients, with a median lead time of 347 days before clinical diagnosis. Our multi-reader study showed that ePAI significantly outperformed 30 board-certified radiologists by 50.3% (P < 0.05) in sensitivity while maintaining a comparable specificity of 95.4% in detecting PDACs early and prediagnostic. These findings suggest its potential of ePAI as an assistive tool to improve early detection of pancreatic cancer.
Large-Scale Label Quality Assessment for Medical Segmentation via a Vision-Language Judge and Synthetic Data
Jan 20, 2026Abstract:Large-scale medical segmentation datasets often combine manual and pseudo-labels of uneven quality, which can compromise training and evaluation. Low-quality labels may hamper performance and make the model training less robust. To address this issue, we propose SegAE (Segmentation Assessment Engine), a lightweight vision-language model (VLM) that automatically predicts label quality across 142 anatomical structures. Trained on over four million image-label pairs with quality scores, SegAE achieves a high correlation coefficient of 0.902 with ground-truth Dice similarity and evaluates a 3D mask in 0.06s. SegAE shows several practical benefits: (I) Our analysis reveals widespread low-quality labeling across public datasets; (II) SegAE improves data efficiency and training performance in active and semi-supervised learning, reducing dataset annotation cost by one-third and quality-checking time by 70% per label. This tool provides a simple and effective solution for quality control in large-scale medical segmentation datasets. The dataset, model weights, and codes are released at https://github.com/Schuture/SegAE.
Auditing Significance, Metric Choice, and Demographic Fairness in Medical AI Challenges
Dec 22, 2025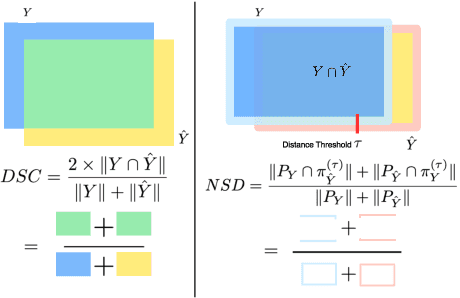
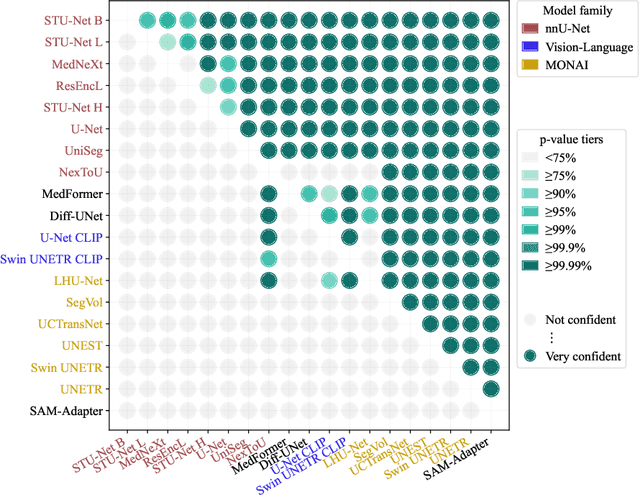
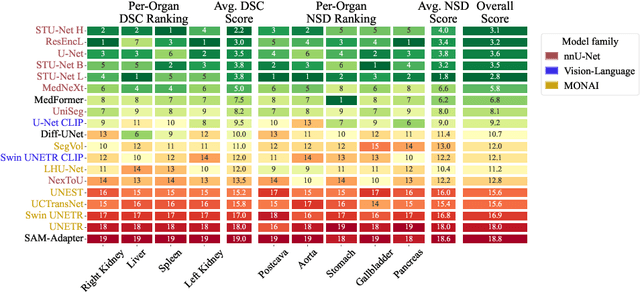
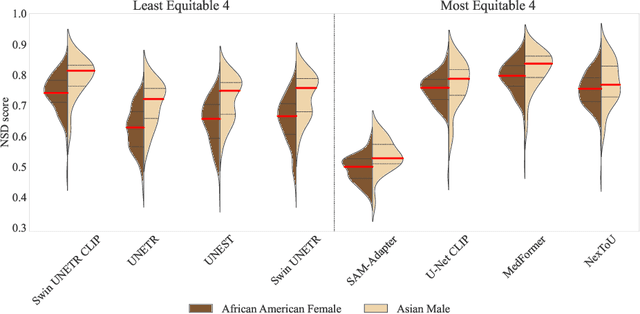
Abstract:Open challenges have become the de facto standard for comparative ranking of medical AI methods. Despite their importance, medical AI leaderboards exhibit three persistent limitations: (1) score gaps are rarely tested for statistical significance, so rank stability is unknown; (2) single averaged metrics are applied to every organ, hiding clinically important boundary errors; (3) performance across intersecting demographics is seldom reported, masking fairness and equity gaps. We introduce RankInsight, an open-source toolkit that seeks to address these limitations. RankInsight (1) computes pair-wise significance maps that show the nnU-Net family outperforms Vision-Language and MONAI submissions with high statistical certainty; (2) recomputes leaderboards with organ-appropriate metrics, reversing the order of the top four models when Dice is replaced by NSD for tubular structures; and (3) audits intersectional fairness, revealing that more than half of the MONAI-based entries have the largest gender-race discrepancy on our proprietary Johns Hopkins Hospital dataset. The RankInsight toolkit is publicly released and can be directly applied to past, ongoing, and future challenges. It enables organizers and participants to publish rankings that are statistically sound, clinically meaningful, and demographically fair.
See More, Change Less: Anatomy-Aware Diffusion for Contrast Enhancement
Dec 08, 2025Abstract:Image enhancement improves visual quality and helps reveal details that are hard to see in the original image. In medical imaging, it can support clinical decision-making, but current models often over-edit. This can distort organs, create false findings, and miss small tumors because these models do not understand anatomy or contrast dynamics. We propose SMILE, an anatomy-aware diffusion model that learns how organs are shaped and how they take up contrast. It enhances only clinically relevant regions while leaving all other areas unchanged. SMILE introduces three key ideas: (1) structure-aware supervision that follows true organ boundaries and contrast patterns; (2) registration-free learning that works directly with unaligned multi-phase CT scans; (3) unified inference that provides fast and consistent enhancement across all contrast phases. Across six external datasets, SMILE outperforms existing methods in image quality (14.2% higher SSIM, 20.6% higher PSNR, 50% better FID) and in clinical usefulness by producing anatomically accurate and diagnostically meaningful images. SMILE also improves cancer detection from non-contrast CT, raising the F1 score by up to 10 percent.
PanTS: The Pancreatic Tumor Segmentation Dataset
Jul 02, 2025Abstract:PanTS is a large-scale, multi-institutional dataset curated to advance research in pancreatic CT analysis. It contains 36,390 CT scans from 145 medical centers, with expert-validated, voxel-wise annotations of over 993,000 anatomical structures, covering pancreatic tumors, pancreas head, body, and tail, and 24 surrounding anatomical structures such as vascular/skeletal structures and abdominal/thoracic organs. Each scan includes metadata such as patient age, sex, diagnosis, contrast phase, in-plane spacing, slice thickness, etc. AI models trained on PanTS achieve significantly better performance in pancreatic tumor detection, localization, and segmentation compared to those trained on existing public datasets. Our analysis indicates that these gains are directly attributable to the 16x larger-scale tumor annotations and indirectly supported by the 24 additional surrounding anatomical structures. As the largest and most comprehensive resource of its kind, PanTS offers a new benchmark for developing and evaluating AI models in pancreatic CT analysis.
Are Vision Language Models Ready for Clinical Diagnosis? A 3D Medical Benchmark for Tumor-centric Visual Question Answering
May 25, 2025Abstract:Vision-Language Models (VLMs) have shown promise in various 2D visual tasks, yet their readiness for 3D clinical diagnosis remains unclear due to stringent demands for recognition precision, reasoning ability, and domain knowledge. To systematically evaluate these dimensions, we present DeepTumorVQA, a diagnostic visual question answering (VQA) benchmark targeting abdominal tumors in CT scans. It comprises 9,262 CT volumes (3.7M slices) from 17 public datasets, with 395K expert-level questions spanning four categories: Recognition, Measurement, Visual Reasoning, and Medical Reasoning. DeepTumorVQA introduces unique challenges, including small tumor detection and clinical reasoning across 3D anatomy. Benchmarking four advanced VLMs (RadFM, M3D, Merlin, CT-CHAT), we find current models perform adequately on measurement tasks but struggle with lesion recognition and reasoning, and are still not meeting clinical needs. Two key insights emerge: (1) large-scale multimodal pretraining plays a crucial role in DeepTumorVQA testing performance, making RadFM stand out among all VLMs. (2) Our dataset exposes critical differences in VLM components, where proper image preprocessing and design of vision modules significantly affect 3D perception. To facilitate medical multimodal research, we have released DeepTumorVQA as a rigorous benchmark: https://github.com/Schuture/DeepTumorVQA.
Text2CT: Towards 3D CT Volume Generation from Free-text Descriptions Using Diffusion Model
May 07, 2025



Abstract:Generating 3D CT volumes from descriptive free-text inputs presents a transformative opportunity in diagnostics and research. In this paper, we introduce Text2CT, a novel approach for synthesizing 3D CT volumes from textual descriptions using the diffusion model. Unlike previous methods that rely on fixed-format text input, Text2CT employs a novel prompt formulation that enables generation from diverse, free-text descriptions. The proposed framework encodes medical text into latent representations and decodes them into high-resolution 3D CT scans, effectively bridging the gap between semantic text inputs and detailed volumetric representations in a unified 3D framework. Our method demonstrates superior performance in preserving anatomical fidelity and capturing intricate structures as described in the input text. Extensive evaluations show that our approach achieves state-of-the-art results, offering promising potential applications in diagnostics, and data augmentation.
MedSegFactory: Text-Guided Generation of Medical Image-Mask Pairs
Apr 09, 2025Abstract:This paper presents MedSegFactory, a versatile medical synthesis framework that generates high-quality paired medical images and segmentation masks across modalities and tasks. It aims to serve as an unlimited data repository, supplying image-mask pairs to enhance existing segmentation tools. The core of MedSegFactory is a dual-stream diffusion model, where one stream synthesizes medical images and the other generates corresponding segmentation masks. To ensure precise alignment between image-mask pairs, we introduce Joint Cross-Attention (JCA), enabling a collaborative denoising paradigm by dynamic cross-conditioning between streams. This bidirectional interaction allows both representations to guide each other's generation, enhancing consistency between generated pairs. MedSegFactory unlocks on-demand generation of paired medical images and segmentation masks through user-defined prompts that specify the target labels, imaging modalities, anatomical regions, and pathological conditions, facilitating scalable and high-quality data generation. This new paradigm of medical image synthesis enables seamless integration into diverse medical imaging workflows, enhancing both efficiency and accuracy. Extensive experiments show that MedSegFactory generates data of superior quality and usability, achieving competitive or state-of-the-art performance in 2D and 3D segmentation tasks while addressing data scarcity and regulatory constraints.
Machine Learning Fleet Efficiency: Analyzing and Optimizing Large-Scale Google TPU Systems with ML Productivity Goodput
Feb 10, 2025



Abstract:Recent years have seen the emergence of machine learning (ML) workloads deployed in warehouse-scale computing (WSC) settings, also known as ML fleets. As the computational demands placed on ML fleets have increased due to the rise of large models and growing demand for ML applications, it has become increasingly critical to measure and improve the efficiency of such systems. However, there is not yet an established methodology to characterize ML fleet performance and identify potential performance optimizations accordingly. This paper presents a large-scale analysis of an ML fleet based on Google's TPUs, introducing a framework to capture fleet-wide efficiency, systematically evaluate performance characteristics, and identify optimization strategies for the fleet. We begin by defining an ML fleet, outlining its components, and analyzing an example Google ML fleet in production comprising thousands of accelerators running diverse workloads. Our study reveals several critical insights: first, ML fleets extend beyond the hardware layer, with model, data, framework, compiler, and scheduling layers significantly impacting performance; second, the heterogeneous nature of ML fleets poses challenges in characterizing individual workload performance; and third, traditional utilization-based metrics prove insufficient for ML fleet characterization. To address these challenges, we present the "ML Productivity Goodput" (MPG) metric to measure ML fleet efficiency. We show how to leverage this metric to characterize the fleet across the ML system stack. We also present methods to identify and optimize performance bottlenecks using MPG, providing strategies for managing warehouse-scale ML systems in general. Lastly, we demonstrate quantitative evaluations from applying these methods to a real ML fleet for internal-facing Google TPU workloads, where we observed tangible improvements.
How Well Do Supervised 3D Models Transfer to Medical Imaging Tasks?
Jan 20, 2025



Abstract:The pre-training and fine-tuning paradigm has become prominent in transfer learning. For example, if the model is pre-trained on ImageNet and then fine-tuned to PASCAL, it can significantly outperform that trained on PASCAL from scratch. While ImageNet pre-training has shown enormous success, it is formed in 2D, and the learned features are for classification tasks; when transferring to more diverse tasks, like 3D image segmentation, its performance is inevitably compromised due to the deviation from the original ImageNet context. A significant challenge lies in the lack of large, annotated 3D datasets rivaling the scale of ImageNet for model pre-training. To overcome this challenge, we make two contributions. Firstly, we construct AbdomenAtlas 1.1 that comprises 9,262 three-dimensional computed tomography (CT) volumes with high-quality, per-voxel annotations of 25 anatomical structures and pseudo annotations of seven tumor types. Secondly, we develop a suite of models that are pre-trained on our AbdomenAtlas 1.1 for transfer learning. Our preliminary analyses indicate that the model trained only with 21 CT volumes, 672 masks, and 40 GPU hours has a transfer learning ability similar to the model trained with 5,050 (unlabeled) CT volumes and 1,152 GPU hours. More importantly, the transfer learning ability of supervised models can further scale up with larger annotated datasets, achieving significantly better performance than preexisting pre-trained models, irrespective of their pre-training methodologies or data sources. We hope this study can facilitate collective efforts in constructing larger 3D medical datasets and more releases of supervised pre-trained models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge